The first week of the academic year at UHasselt has come to an end, while colleagues at UGent and KULeuven are still preparing for the start of their academic year next week. Good luck to all of you.
This week started full throttle for me, with classes for each of my six courses. After introductions in classes with new students (for me) in the second bachelor chemistry and first master materiomics, and a general overview in the different courses, we quickly dove into the subject at hand. 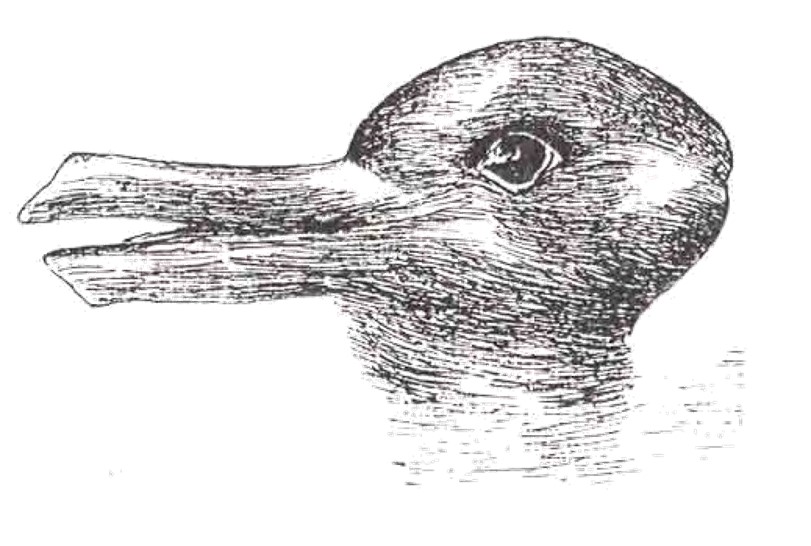
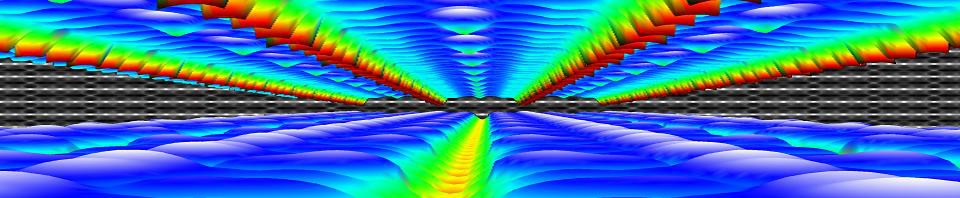
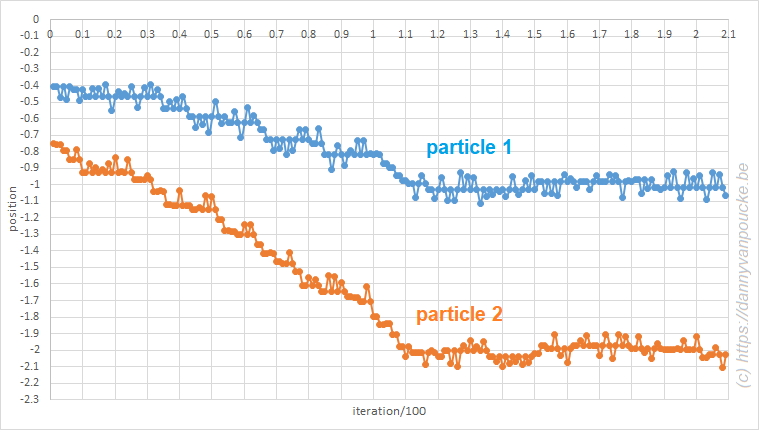
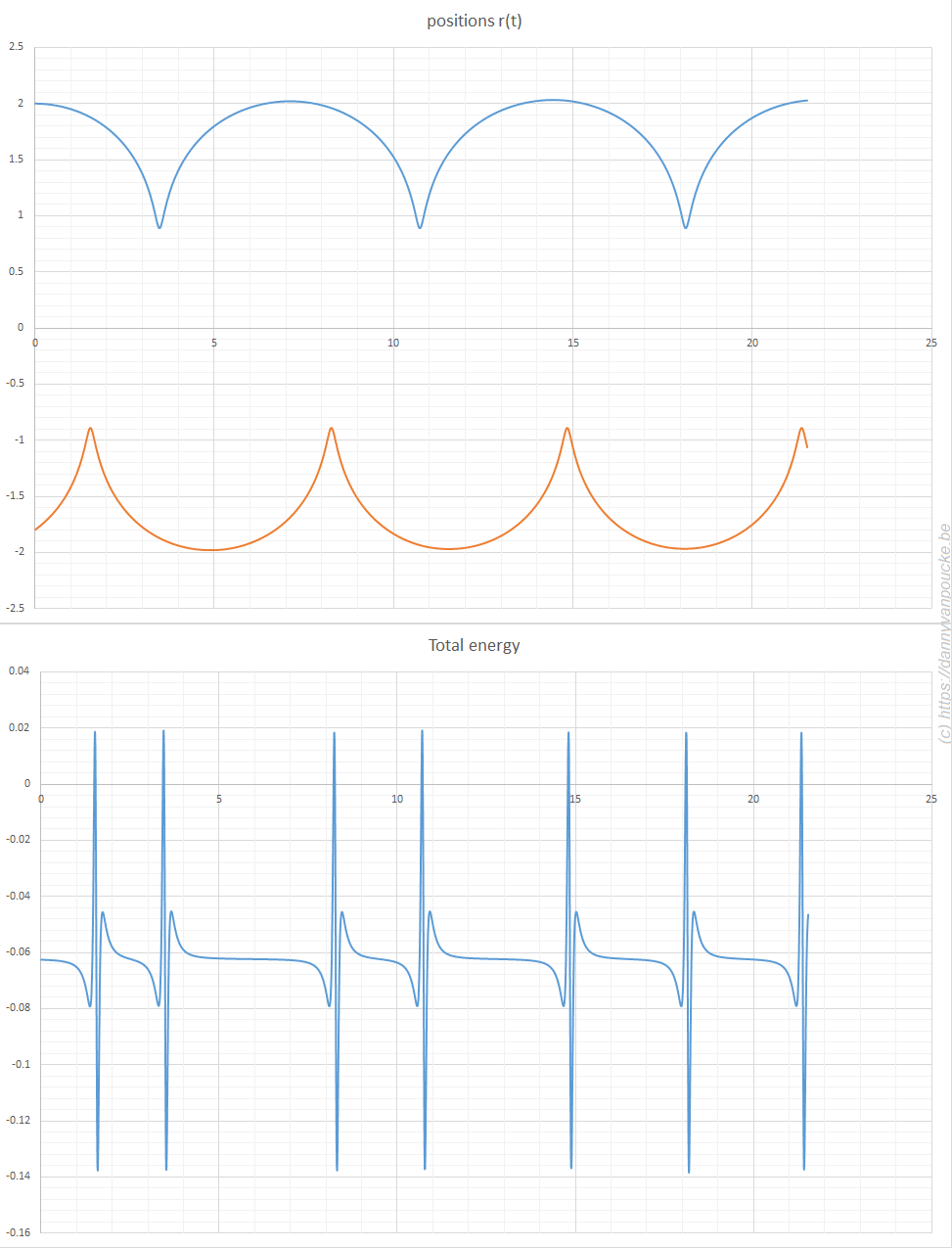
The second bachelor students (introduction to quantum chemistry) got a soft introduction into (some of) the historical events leading up to the birth of quantum mechanics such as the black body radiation, the atomic model and the nature of light. They encountered the duck-rabbit of particle-wave duality and awakened their basic math skills with the standing wave problem. For the third bachelor students, the course on quantum and computational chemistry started with a quick recap of the course introduction to quantum mechanics, making sure they are all again up to speed with concepts like braket-notation and commutator relations.
For the master materiomics it was also a busy week. We kicked of the 1st Ma course Fundamentals of materials modeling, which starts of calm and easy with a general picture of the role of computational research as third research paradigm. We discussed in which fields computational research can be found (flabbergasting students with an example in Theology: a collaboration between Sylvia Wenmackers & Helen De Cruz), approximation vs idealization, examples of materials research at different scales, etc. As a homework assignment the students were introduced into the world of algorithms through the lecture of Hannah Fry (Should computers run the world). For the 2nd Ma, the courses on Density Functional Theory and Machine learning and artificial intelligence in modern materials science both started. The lecture of the former focused on the nuclear wave function and how we (don’t) deal with it in DFT, but still succeed in optimizing structures. During the lecture on AI we dove into the topics of regularization and learning curves, and extended on different types of ensemble models.
At the end of week 1, this brings me to a total of 12h of lectures. Upwards and onward to week 2.