MSc Thesis presentation of Brent Motmans and Eleonora Thomas (master materiomics students 2025). Both presenting applications of ML in materials research: Machine Learning particle sizes using small lab-scale datasets (Brent) and development of Machine Learned Interatomic Potentials for the modelling of (the dynamics of) H-based defects in diamond.
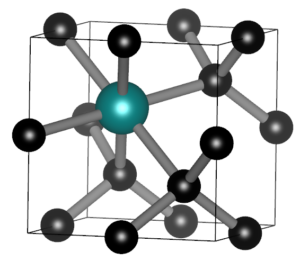
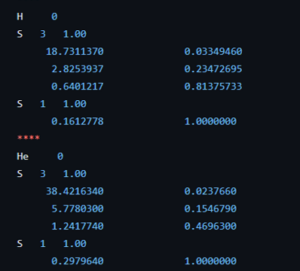
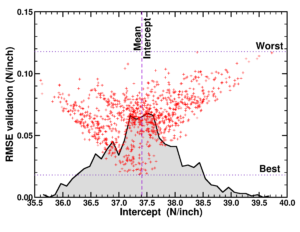
Today we had the MSc presentations of the master Materiomics. The culmination of two year of hard study and intens research activities resulting in a final master thesis paper. This year the QuATOMs group hosted two MSc students: Brent Motmans and Eleonora Thomas. Brent Motmans performed his research in a collaboration between the QuATOMs and DESINe groups, and investigated the application of small data machine learning for the prediction of the particle size of Cu nanoparticles. His study shows that even with a dataset of less than 20 samples a reasonable 6 feature model can be created. As in previous research, he found that standard hyperparameter tuning fails, but human intervention can resolve this issue. Eleonora Thomas on the other hand introduced Machine Learned Interatomic Potentials (MLIPs) into the group. By investigating different in literature available MLIPs, she pinpointed strengths and weaknesses of the different models, as well as the technical needs for persuing such research further in our group. As collatoral, she was able to generate a model for H diffusion in diamond, with an MAE for the total energy of <10meV/atom, competing with models like google deep-mind’s GNoME.
While working on their MSc thesis, Brent and Eleonora also applied for fellowship funding for a PhD position, and we are happy to announce both Brent and Eleonora won their grant, and will be starting in the QuATOMs group as new PhD students comming academic year. Eleonora Thomas will be working on the modelling of Lignin solvation, while Brent will work in a collaboration with the HyMAD group on the modeling of hybrid perovskites.


 During the last year,
During the last year,