After week four, this fifth week of the academic year is most arguably the most intense and hectic week of teaching. With 22h of classes and still two classes that needed to be prepared from scratch (even including weekends time was running out), I’m tired but happy it is over. However, all the effort is worth it, and I was happy to hear the students of our materiomics program at UHasselt appreciate the effort put into creating their classes, during an evaluation meeting.
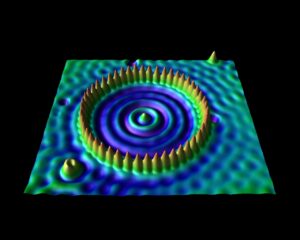
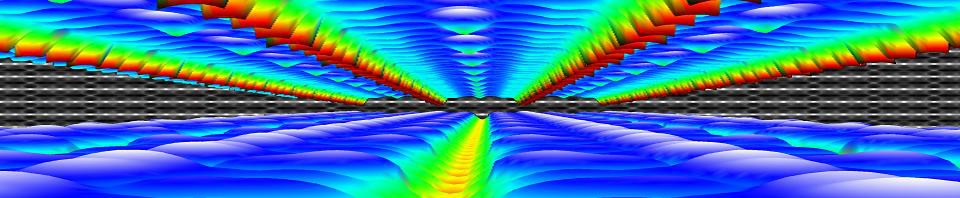
The corral is an artificial structure created from 48 iron atoms (the sharp peaks) on a copper surface. The wave patterns in this scanning tunneling microscope image are formed by copper electrons confined by the iron atoms. Don Eigler and colleagues created this structure in 1993 by using the tip of a low-temperature scanning tunneling microscope (STM) to position iron atoms on a copper surface, creating an electron-trapping barrier. This was the first successful attempt at manipulating individual atoms and led to the development of new techniques for nanoscale construction.
source: https://www.nisenet.org/catalog/scientific-image-quantum-corral-top-view
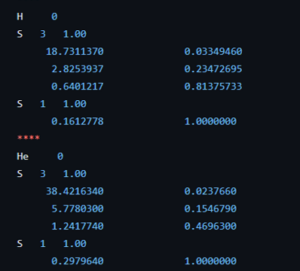
For the second bachelor students in chemistry the introduction to quantum chemistry finally put them into contact with some “real” quantum mechanics when they were introduced into the world of potential barriers, steps, and wells. Though these are still abstract and toy-model in nature, they provide a first connection with reality, where they can be seen as crude approximations of the potential experienced by valence electrons near the surface, or STM experiments. They were also introduced to my favorite quantum system related to this course: the quantum corral. Without any effort it can be used in half a dozen situations with varying complexity to show and learn basic quantum mechanics. For the third bachelor chemistry students the course quantum and computational chemistry finally provided them the long promised first example of a non-trivial quantum chemical object: The Helium atom. With it’s two electrons, we break free of the H-atom(-like) world. Using perturbation theory and Slater determinant wave functions, we made our first approximations of its energy. In addition, these students also had a seminar for their course Introductory lectures in preparation to the bachelor project (Kennismakingstraject m.b.t. stage en eindproject, in Dutch). During this lecture I gave a brief introduction and overview of the work I did in the past and the work we do in our research group QuATOMs, which although “quantum” is quite different of what the students experience during their courses on quantum chemistry.
In the materiomics program, the first master students continued their travels into the basics of force-fields during the lecture of the course Fundamentals of materials modelling. The exercise class of this week upped the ante by moving from ASE to LAMMPS for practical modeling of alkane chains, which was also the topic of their second lab session. In the course Properties of functional materials, we investigated the ab initio modelling of vibrations. During the exercise classes we investigated precalculated phonon spectra in the materials-project database, as well as calculated our own vibrational spectrum at the gamma-point of the first Brillouin zone. During the second master course Machine learning and artificial intelligence in modern materials science the central theme was GIGO (Garbage-In-Garbage-Out). How can we make sure our data is suitable and good enough for our models to return useful results. We therefore looked into data-preparation & cleaning, as well as clustering methods.
At the end of this week, we have added another 22h of live lectures and ~1h of video lectures, putting our semester total at 74h of live lectures. Upwards and onward to week 6.