In the previous sessions of this tutorial on Object Oriented Programming in Fortran 2003, the basics of OO programming, including the implementation of constructors and destructors as well as operator overloading were covered. The resulting classes have already become quite extended (cf. github source). Although at this point it is still very clear what each part does and why certain choices were made, memory fades. One year from now, when you revisit your work, this will no longer be the case. Alternately, when sharing code, you don’t want to have to dig through every line of code to figure out how to use it. These are just some of the reasons why code documentation is important. This is a universal habit of programming which should be adopted irrespective of the programming-language and-paradigm, or size of the code base (yes, even small functions should be documented).
In Fortran, comments can be included in a very simple fashion: everything following the “!” symbol (when not used in a string) is considered a comment, and thus ignored by the compiler. This allows for quick and easy documentation of your code, and can be sufficient for single functions. However, when dealing with larger projects retaining a global overview and keeping track of interdependencies becomes harder. This is where automatic documentation generation software comes into play. These tools parse specifically formatted comments to construct API documentation and user-guides. Over the years, several useful tools have been developed for the Fortran language directly, or as a plugin/extension to a more general tool:
- ROBODoc : A tool capable of generating documentation (many different formats) for any programming/script language which has comments. The latest update dates from 2015.
- Doctran : This tool is specifically aimed at free-format (≥ .f90 ) fortran, and notes explicitly the aim to deal with object oriented f2003. It only generates html documentation, and is currently proprietary with license costs of 30£ per plugin. Latest update 2016.
- SphinxFortran : This extension to SphinxFortran generates automatic documentation for f90 source (no OO fortran) and generates an html manual. This package is written in python and requires you to construct your config file in python as well.
- f90doc / f90tohtml : Two tools written in Perl, which transform f90 code into html webpages.
- FotranDOC : This tool (written in Fortran itself) aims to generate documentation for f95 code, preferably in a single file, in latex. It has a simple GUI interface, and the source of the tool itself is an example of how the fortran code should be documented. How nice is that?
- FORD : Ford is a documentation tool written in python, aimed at modern fortran (i.e. ≥ f90).
- Doxygen : A multi-platform automatic documentation tool developed for C++, but extended to many other languages including fortran. It is very flexible, and easy to use and can produce documentation in html, pdf, man-pages, rtf,… out of the box.
As you can see, there is a lot to choose from, all with their own quirks and features. One unfortunate aspect is the fact that most of these tools use different formatting conventions, so switching from one to the another is not an exercise to perform lightly. In this tutorial, the doxygen tool is used, as it provides a wide range of options, is multi-platform, supports multiple languages and multiple output formats.
As you might already expect, Object Oriented Fortran (f2003) is a bit more complicated to document than procedural Fortran, but with some ingenuity doxygen can be made to provide nice documentation even in this case.
1. Configuring Doxygen
Before you can start you will need to install doxygen:
- Go the the doxygen-download page and find the distribution which is right for you (Windows-users: there are binary installers, no hassle with compilations 🙂 ).
- Follow the installation instructions, also install GraphViz, this will allow you to create nicer graphics using the dot-tool.
- Also get a pdf version of the manual (doxygen has a huge number of options)
With a nicely installed doxygen, you can make use of the GUI to setup a configuration suited to your specific needs and generate the documentation for your code automatically. For Object Oriented Fortran there are some specific settings you should consider: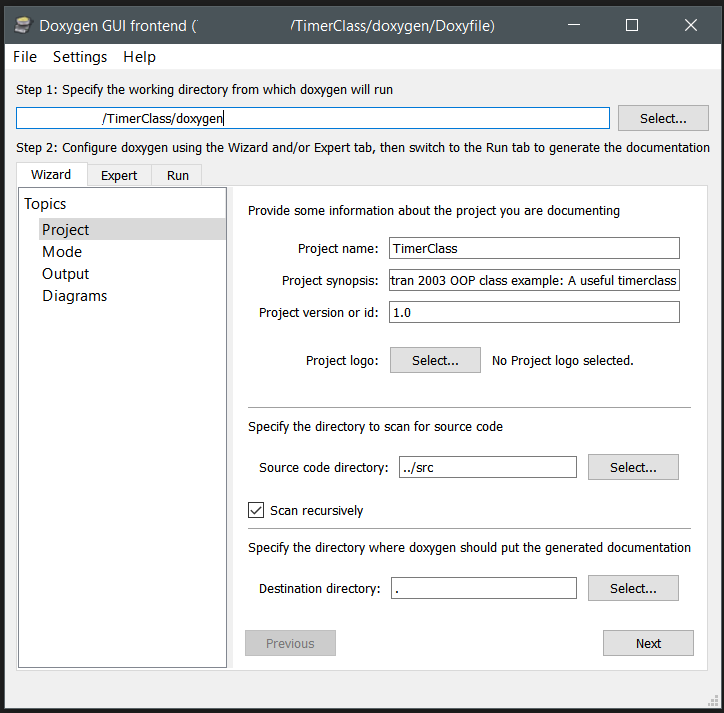
-
Wizard tab
- Project Topic : Fill out the different fields. In a multi-file project, with source stored in a folder structure, don’t forget to select the tick-box “Scan recursively” .
- Mode Topic : Select “Optimize for Fortran output”.
- Output Topic : Select one or more output formats you wish to generate: html, Latex (pdf), map-pages, RTF, and XML
- Diagrams Topic: Select which types of diagrams you want to generate.
-
Expert tab
(Provides access each single configuration option to set in doxygen, so I will only highlight a few. Look through them to get a better idea of the capabilities of doxygen.)
- Project Topic :
- EXTENSION_MAPPING: You will have to tell doxygen which fortran extensions you are using by adding them, and identifying it as free format fortran: e.g. f03=FortranFree (If you are also including text-files to provide additional documentation, it is best to add them here as well as free format fortran).
- Build Topic:
- CASE_SENSE_NAMES: Even though Fortran itself is not case sensitive, it may be nice to keep the type of casing you use in your code in your documentation. Note, however, that even though the output may have upper-case names, the documentation itself will require lower-case names in references.
- Messages Topic:
- WARN_NO_PARAMDOC: Throw a warning if documentation is missing for a function variable. This is useful to make sure you have a complete documentation.
- Source Browser Topic:
- SOURCE_BROWSER: Complete source files are included in the documentation.
- INLINE_SOURCES: Place the source body with each function directly in the documentation.
- HTML Topic:
- FORMULA_FONTSIZE: The fontsize used for generated formulas. If 10 pts is too small to get a nice effect of formulas embedded in text.
- Dot Topic:
- HAVE_DOT & DOT_PATH: If you installed GraphViz
- DOT_GRAPH_MAX_NODES: Maximum number of nodes to draw in a relation graph. In case of larger projects, 50 may be too small.
- CALL_GRAPH & CALLER_GRAPH: Types of relation graphs to include.
-
Run tab
- Press “Run doxygen” and watch how your documentation is being generated. For larger projects this may take some time. Fortunately, graphics are not generated anew if they are present from a previous run, speeding things up. (NOTE: If you want to generate new graphics (and equations with larger font size), make sure to delete the old versions first.) Any warnings and errors are also shown in the main window.
- Once doxygen was run successfully, pressing the button “Show HTML output” will open a browser and take you to the HTML version of the documentation.
Once you have a working configuration for doxygen, you can save this for later use. Doxygen allows you to load an old configuration file and run immediately. The configuration file for the Timer-class project is included in the docs folder, together with the pdf-latex version of the generated documentation. Doxygen generates all latex files required for generating the pdf. To generate the actual pdf, a make.bat file needs to be run (i.e. double-click the file, and watch it run) in a Windows environment.
2. Documenting Fortran (procedural)
Let us start with some basics for documenting Fortran code in a way suitable for doxygen. Since doxygen has a very extensive set of options and features, not all of them can be covered. However, the manual of more than 300 pages provides all the information you may need.
With doxygen, you are able to document more or less any part of your code: entire files, modules, functions or variables. In each case, a similar approach can be taken. Let’s consider the documentation of the TimeClass module:
!++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
!> \brief The <b>TimeClass module</b> contains the
!! \link ttime TTime class\endlink used by the
!! \link timerclass::ttimer TTimer class\endlink for practical timing.
!!
!! @author Dr. Dr. Danny E. P. Vanpoucke
!! @version 2.0-3 (upgrades deprecated timing module)
!! @date 19-03-2020
!! @copyright https://dannyvanpoucke.be
!!
!! @warning Internally, Julian Day Numbers are used to compare dates. As a
!! result, *negative* dates are not accepted. If such dates are created
!! (*e.g.*, due to a subtraction), then the date is set to zero.
!!
!! This module makes use of:
!! - nothing; this module is fully independent
!<-----------------------------------------------------------------------
module TimeClass
implicit none
private
The documentation is placed in a standard single or multi-line fortran comment. In case of multi-line documentation, I have the personal habit turning it into a kind of banner starting with a “!+++++++++” line and closing with a “!<——————-” line. Such choices are your own, and are not necessary for doxygen documentation. For doxygen, a multi-line documentation block starts with “!>” and ends with “!<“ . The documentation lines in between can be indicated with “!!”. This is specifically for fortran documentation in doxygen. C/C++ and other languages will have slightly different conventions, related to their comment section conventions.
In the block above, you immediately see certain words are preceded by an “@”-symbol or a “\”, this indicates these are special keywords. Both the “@” and “\” can be used interchangeably for most keywords, the preference is again personal taste. Furthermore, doxygen supports both html and markdown notation for formatting, providing a lot of flexibility. The multi-line documentation is placed before the object being documented (here an entire module).
Some keywords:
- \brief : Here you can place a short description of the object. This description is shown in parts of the documentation that provide an overview. Note that this is also the first part of the full documentation of the object itself. After a blank line, the \details(this keyword does not need to provided explicitly) section starts, providing further details on the object. This information is only visible in the documentation of the object itself.
- \link … \endlink, or \ref : These are two option to build links between parts of your documentation. You can either use \ref nameobject or \link nameobject FormattedNameObject \endlink. Note that for fortran, doxygen uses an all non-capitalized namespace, so YourObject needs to be referenced as \ref yourobject or you will end up with an error and a missing link. So if you want your documentation to show YourObject as a link instead of yourobject, you can use the \link … \endlink construction.
- “::” : Referring to an element of an object can be done by linking the element and the object via two colons: object::element . Here it is important to remember that your module is an object, so linking to an element of a module from outside that module requires you to refer to it in this way.
- @author : Provide information on the author.
- @version : Provide version information.
- @date : Provide information on the date.
- @copyright : Provide information on the copyright.
- @warning : Provides a highlighted section with warning information for the user of your code (e.g., function kills the program when something goes wrong).
- @todo : [not shown] If you still have some things to do with regard to this object you can use this keyword. More interestingly, doxygen will also create a page where all to-do’s of the entire project are gathered, and link back to the specific code fragments.
!++++++++++++++++++++++++++++++++++++++++++++++
!>\brief Function to subtract two \link ttime TTime\endlink instance
!! via the "-" operator. This is the function
!! performing the actual operator overloading.
!!
!! \b usage:
!! \code{.f03}
!! Total = this - that
!! \endcode
!! This line also calls the \link copy assignment operator\endlink.
!!
!! \note The result should remain a positive number.
!!
!! @param[in] this The \link ttime TTime\endlink instance before
!! the "-" operator.
!! @param[in] that The \link ttime TTime\endlink instance after
!! the "-" operator.
!! \return Total The \link ttime TTime\endlink instance representing
!! the difference.
!<---------------------------------------------
pure function subtract(this,that) Result(Total)
class(TTime), intent(in) :: this, that
Type(TTime) :: total
When documenting functions and subroutines there are some addition must-have keywords.
- @param[in] , @param[out] ,or@param[in,out] : Provide a description for each of the function parameters, including their intent: “in”, “out”, or “in,out” (note the comma!).
- \return : Provides information on the return value of the function.
- \b, \i : The next word is bold or italic
- \n : Start a newline, without starting a new paragraph.
- \note : Add a special note in your documentation. This section will be high lighted in a fashion similar to @warning.
- \code{.f03}…\endcode : This environment allows you to have syntax highlighted code in your documentation. The language can be indicated via the “extension” typical for said language. In this case: fortran-2003.
- \f$ … \f$, or \f[ … \f] : Sometimes equations are just that much easier to convey your message. Doxygen also supports latex formatting for equations. These tags can be used to enter a latex $…$ or \[ \] math environments. The equations are transformed into small png images upon documentation generation, to be included in the html of your documentation. There are two important aspects to consider when using this option:
- Font size of the equation: Check if this is sufficient and don’t be afraid to change the font size to improve readability.
- Compilation is not halted upon an error: If the latex compiler encounters an error in your formula it just tries to continue. In case of failure, the end result may be missing or wrong. Debugging latex equations in doxygen documentation can be quite challenging as a result. So if you are using large complex equations, it may be advised to run them in a pure latex environment, and only past them in the documentation once you are satisfied with the result.
3. Documenting Fortran Classes
With the knowledge of the previous section, it is relatively easy to document most fortran code. Also the type of object orientation available in fortran 95, in which a fortran module is refurbished as a class. True fortran classes in contrast tend to give a few unexpected issues to deal with. Lets have a look at the documentation of the TTime class of the TimeClass module:
!+++++++++++++++++++++++++++++++++++++++
!> @class ttime
!! \brief The TTime class contains all time functionality
!! with regard to a single time stamp.
!<-------------------------------------
type, public :: TTime
private
integer :: year !< @private The year
integer :: month !< @private The month (as integer).
...
contains
private
procedure, pass(this),public :: SetTime !< @copydoc timeclass::settime
procedure, pass(this) :: CalculateJDN !< @private @copydoc timeclass::calculatejdn
procedure, pass(this) :: SetJDN !< @private @copydoc timeclass::setjdn
...
procedure, pass(this) :: copy !< @private @copydoc timeclass::copy
...
generic, public :: assignment(=) => copy !< @copydoc timeclass::copy
!> @{ @protected
final :: destructor !< @copydoc timeclass::destructor
!> @}
end type TTime
! This is the only way a constructor can be created,
! as no "initial" exists, emulates the C++ constructor behavior
interface TTime
module procedure constructor
end interface TTime
To make sure doxygen generates a class-like documentation for our fortran class, it needs to be told it is a class. This can be done by documenting the class itself and using the keyword @class nameclass, with nameclass the name doxygen will use for this class (so you can choose something different from the actual class name). Unfortunately, doxygen will call this a “module” in the documentation (just poor luck in nomenclature). On the module page for the ttime class a listing is provided of all elements given in the class definition. The documentation added to each member (e.g.,:
integer :: year !< @private The year
is shown as “\brief” documentation. By default all members of our function are considered as public. Adding the @private, @public, or @protected keyword instructs doxygen explicitly to consider these members as private, public or protected. (I used protected in the ttime code not as it should be used in fortran, but as a means of indicating the special status of the final subroutine (i.e. protected in a C++ way).)
However, there seems to be something strange going on. When following the links in the documentation, we do not end up with the documentation provided for the functions/subroutines in the body of our timeclass module. Doxygen seems to consider these two distinct things. The easiest way to link the correct information is by using the keyword @copydoc functionreference . The documentation is (according to doxygen) still for two distinctly different objects, however, this time they have the exact same documentation (unless you add more text on the member documentation line). In this context, it interesting to know there is also @copybrief and @copydetails which can be used to only copy the brief/details section.
In this example, the constructor interface is not documented, as this created confusion in the final documentation since doxygen created a second ttime module/object linked to this interface. However, not documenting this specific instance of the constructor does not create such a large issue, as the module(the fortran module) function itself is documented already.
Conclusion
Documenting fortran classes can be done quite nicely with doxygen. It provides various modes of output: from a fully working website with in-site search engine to a hyperlinked pdf or RTF document. The flexibility and large number of options may be a bit daunting at first, but you can start simple, and work your way up.
As Fortran is supported as an extension, you will need to play around with the various options to find which combination gives the effect you intended. This is an aspect present in all automated code documentation generation tools, since object oriented Fortran is not that widely used. Nonetheless, doxygen provides a very powerful tool worth your time and effort.
PS: